Luna L. Sánchez Reyes
Postdoctoral Research Scholar
University of California, Merced
Hello!
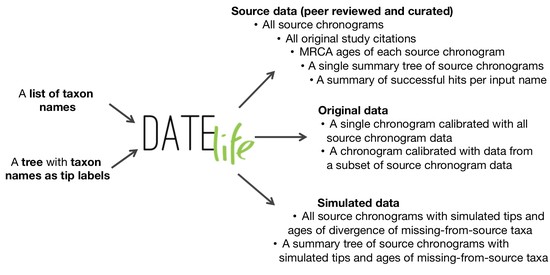
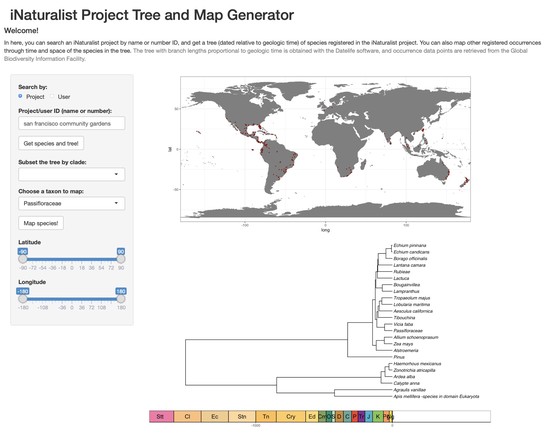
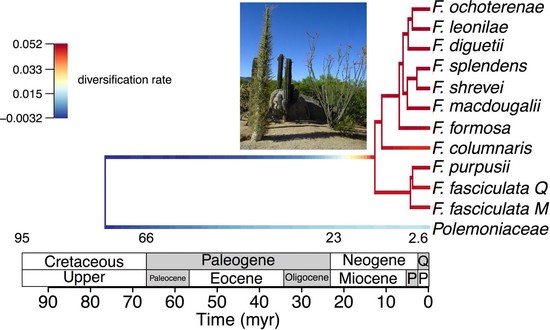
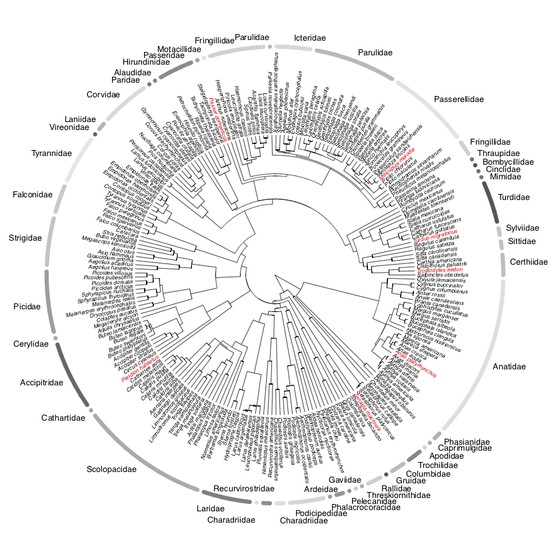
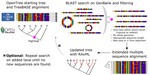
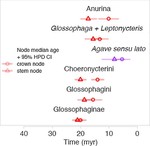
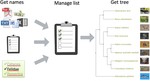
I am a biologist specialized in evolutionary biology. I am currently working as a postdoc at the McTavish Lab, at the School of Natural Sciences of the University of California, Merced. My research interests include species diversification processes, timing of species origin, the intercept between micro and macroevolution, open science, and science communication and reproducibility. For the past few years, I have been working on method and software development for various projects, including platforms such as Datelife, Phylotastic, and most recently Open Tree of Life.
Interests
- Effects of statistical uncertainty on evolutionary inferences
- Coding with R and Python
- Open Science in Education and Research
Education
-
Biological Sciences PhD, 2016
Universidad Nacional Autónoma de México
-
Biology BSc, 2010
Universidad Nacional Autónoma de México